Gender dimorphism in early Alzheimer’s pathology: Brain connectivity with MRI and behavioral analysis in a mouse humanized for the genes of App and MAPT
Co-directed Interdisciplinary PhD project – ICube Lab / LNCA;
University of Strasbourg / CNRS Project funded by FRM (Fondation pour la Recherche Medicale)
RESEARCH PROJECT:
Women have a higher rate of cognitive decline than men at a preclinical stage of Alzheimer’s disease (AD). Moreover – during the AD progression, cognitive decline, brain atrophy and tau pathology, are more pronounced in women (1). Finding gender-specific biomarkers that predict AD onset and evolution is absolutely critical for accurate diagnosis and personalized therapies (2).
AIM and approaches: In this project we will combine functional and structural brain Magnetic Resonance Imaging (MRI), network analysis approaches and behavioral phenotyping to investigate the gender specific connectivity signatures of AD.
We will use in a longitudinal design a new preclinical model – the AppNL-F/MAPT double knock-in (dKI) mouse; humanized for the genes of App and MAPT (3). In this model, LNCA (Laboratory of Cognitive and Adaptive Neurosciences) recently identified an early vulnerability of the females for cognitive deficits; however the circuitries involved in this early phenotype are not known. Meanwhile, in another model of AD, a tauopathy mouse model – the ICube/LNCA labs recently demonstrated using resting state functional MRI (rsfMRI) that remodeling of brain networks architecture precede behavioral deficits, and moreover can highlight compensatory pathways
(4).
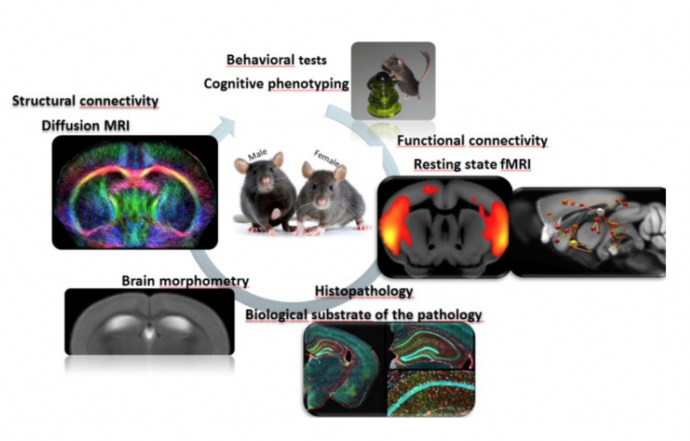
Based on these findings we will apply multi-variate analysis approaches: (i) resting state functional MRI to characterize the dynamics of functional network architecture and to identify sex specific network signatures and networks’ hubs, critical for memory deficits occurring overtime. Open-ended (whole brain) (5, 6) and hypothesis-driven analyses will be developed to elucidate circuits underlying the pathology. (ii) High angular resolution diffusion imaging (HARDI) and fiber tractography (7) to explore whole brain microstructure and the dynamics of fiber density alterations at different time points over life span. Brain tractography will be associated to anatomical imaging for brain morphometry. (iii) Behavioral tests and histopathological analysis (LNCA expertise) to characterize the cognitive phenotype.
CONTEXT: The imaging will be carried-out within the “I
ntegrative Multi-modal Imaging in Healthcare – IMIS” Team, led by Laura-Adela Harsan at ICube (https://icube.unistra.fr/equipes/, Strasbourg). The IMIS team includes experts in MR techniques and signal modeling, brain networks analysis, neurobiology and preclinical models of brain disorders. The project will use the ICube lab imaging platform facilities (7T MRI animal scanner; bioluminescence and microscopy tools). The expertise in animal behavior and histopathology will be provided via co-supervision with Chantal Mathis, leading the ENGRAM team at LNCA.
CANDIDATE PROFILE: The candidate should have background in Neuroscience and/or MR technologies and MR data processing; and be highly motivate
d to work within an interdisciplinary context for optimizing/validating/applying MR methodology in preclinical environment. The selected candidate should have knowledge regarding brain anatomy and function and animal physiology. Programming skills (MATLAB, Phyton) for MRI data processing in correlation with behavioral results are appreciated, as well as good track record and good proficiency in english.
CONTACT: Laura-Adela Harsan (harsan@unistra.fr); Engineering science, computer science and imaging laboratory
and Chantal Mathis (chantal.mathis@unistra.fr), Laboratory of Cognitive and Adaptive Neurosciences
Refs: (1) Duarte-Guterman et al, bioRxiv, online Aug. 23, 2019; (2) Ferretti et al, Nat Rev Neurol 14:457, 2018; (3) Saito et al, J Biol Chem 294:12754, 2019 ; (4) Degiorgis L et al., Brain, In press; (5) Mechling et al, PNAS 113:11603, 2016; (6) Arefin et al, Brain Connect 7:526, 2017; (7) Harsan et al, PNAS 110: E1797, 2013